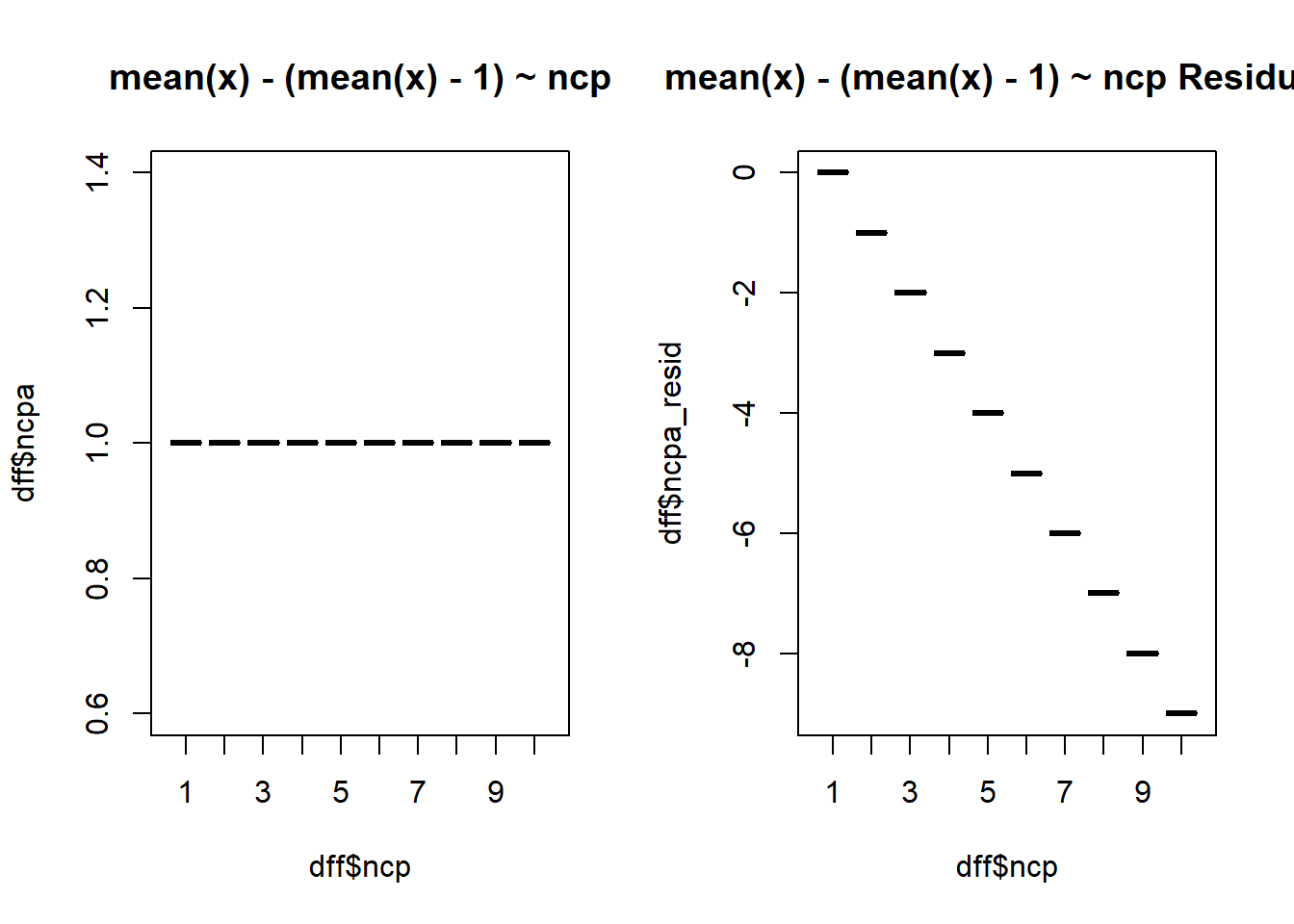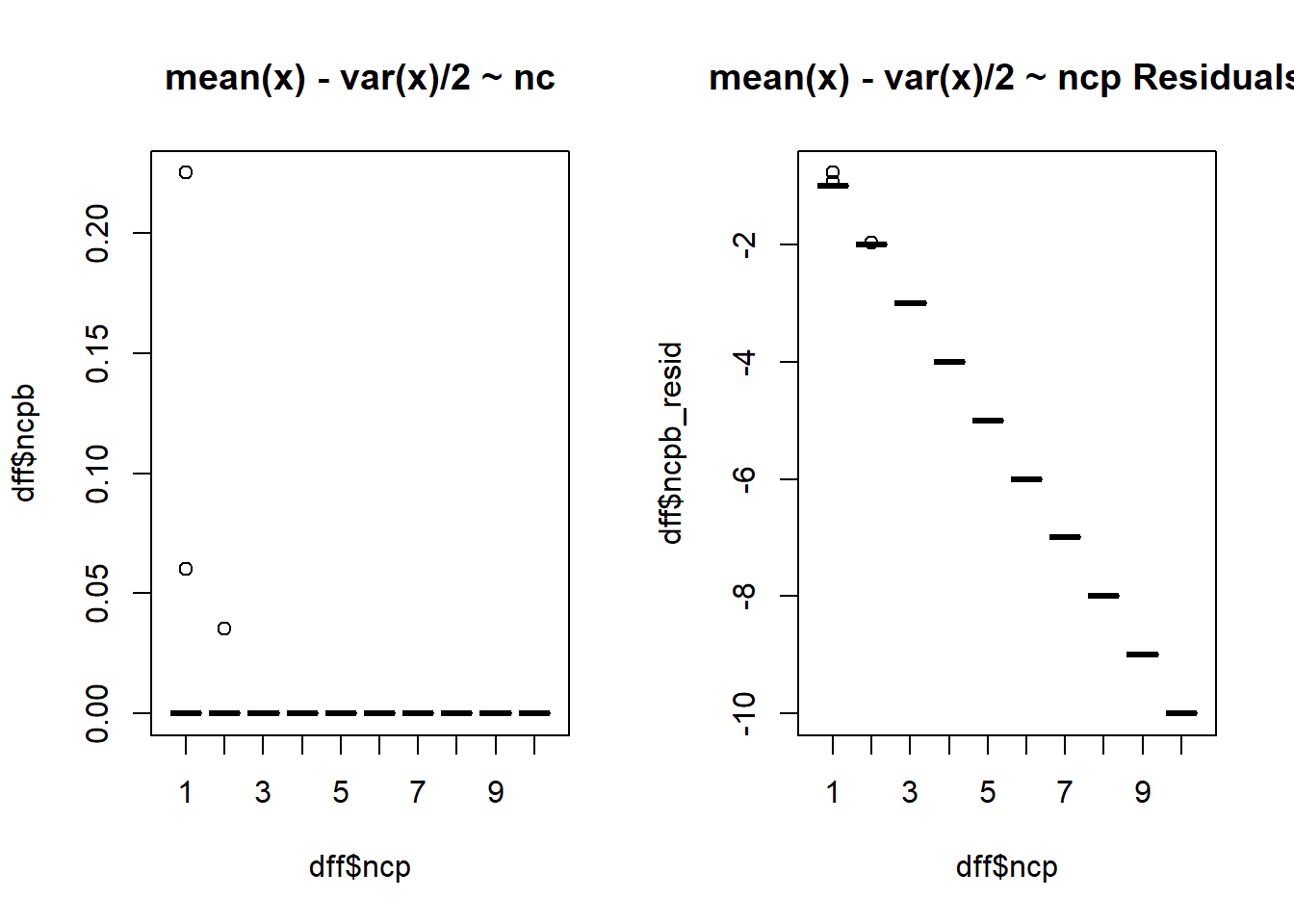Rows: 987
Columns: 19
$ n <dbl> 284, 284, 284, 284, 284, 284, 284, 284, 284, 284, 284, 284,…
$ df <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,…
$ ncp <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 1, 2, 3, 4, 5, 6, 7, 8, 9, 1…
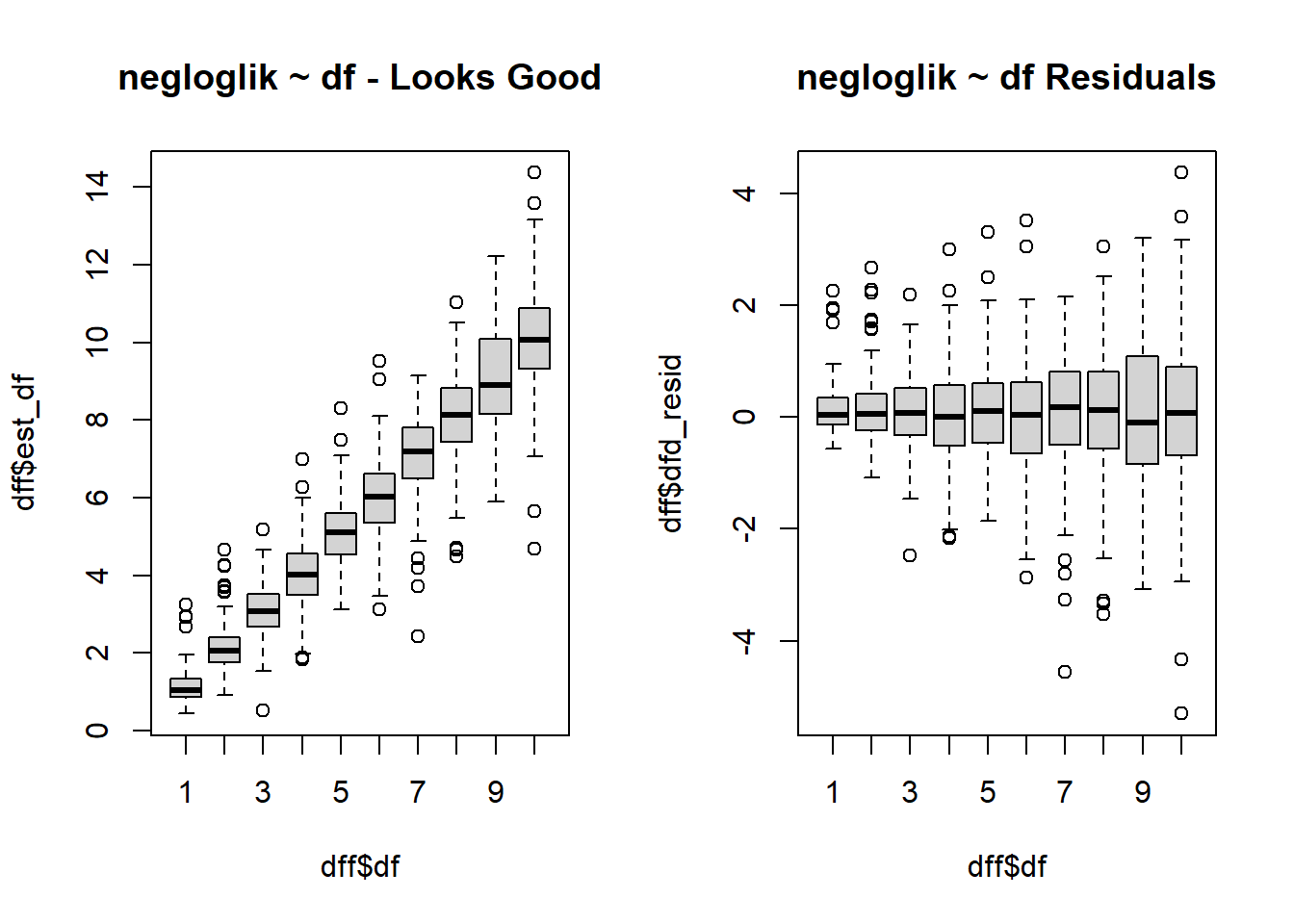
$ est_df <dbl> 1.1770904, 0.9905994, 0.9792179, 0.7781877, 1.5161669, 0.82…
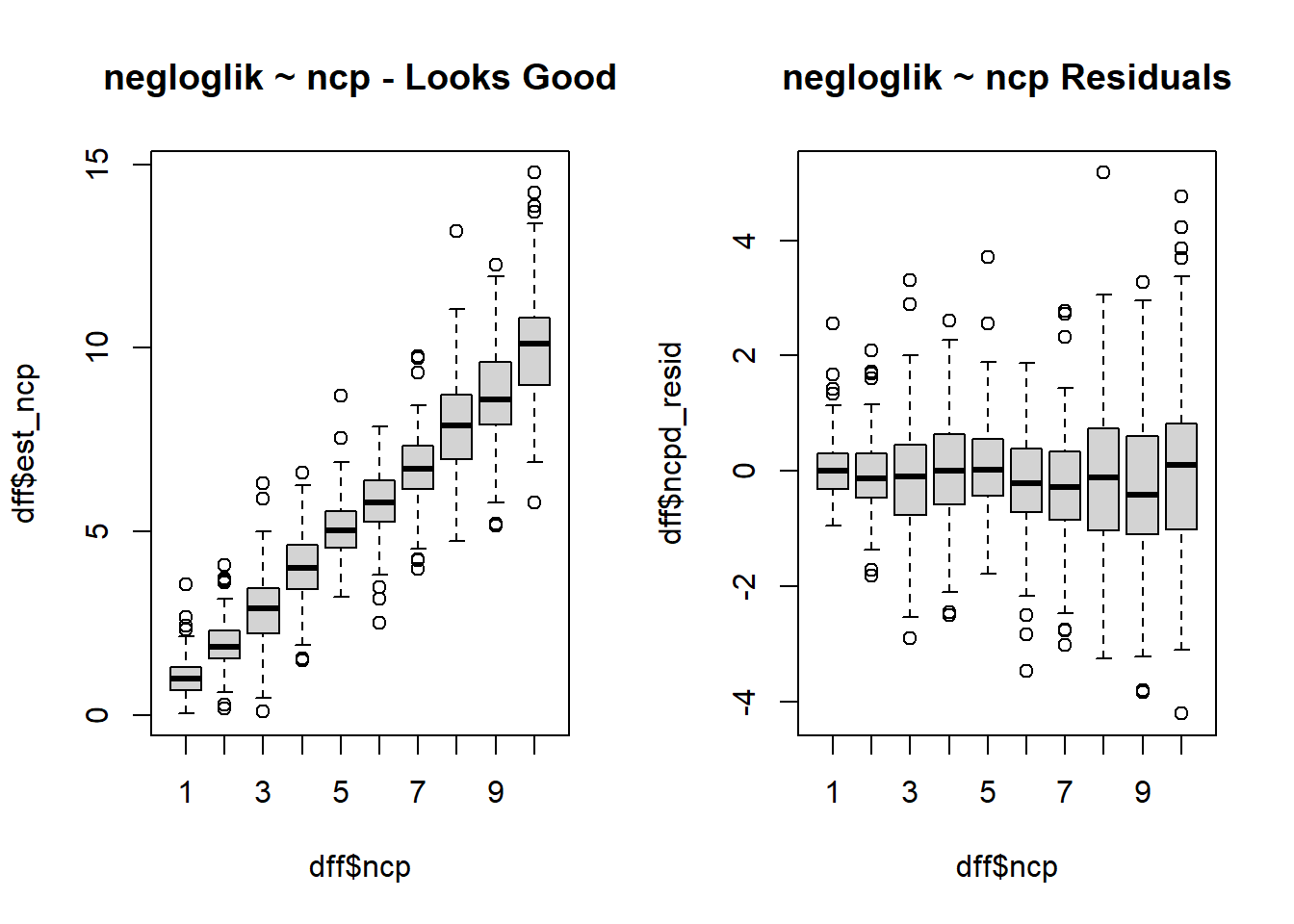
$ est_ncp <dbl> 0.7231638, 1.9462325, 3.0371756, 4.2347494, 3.7611119, 6.26…
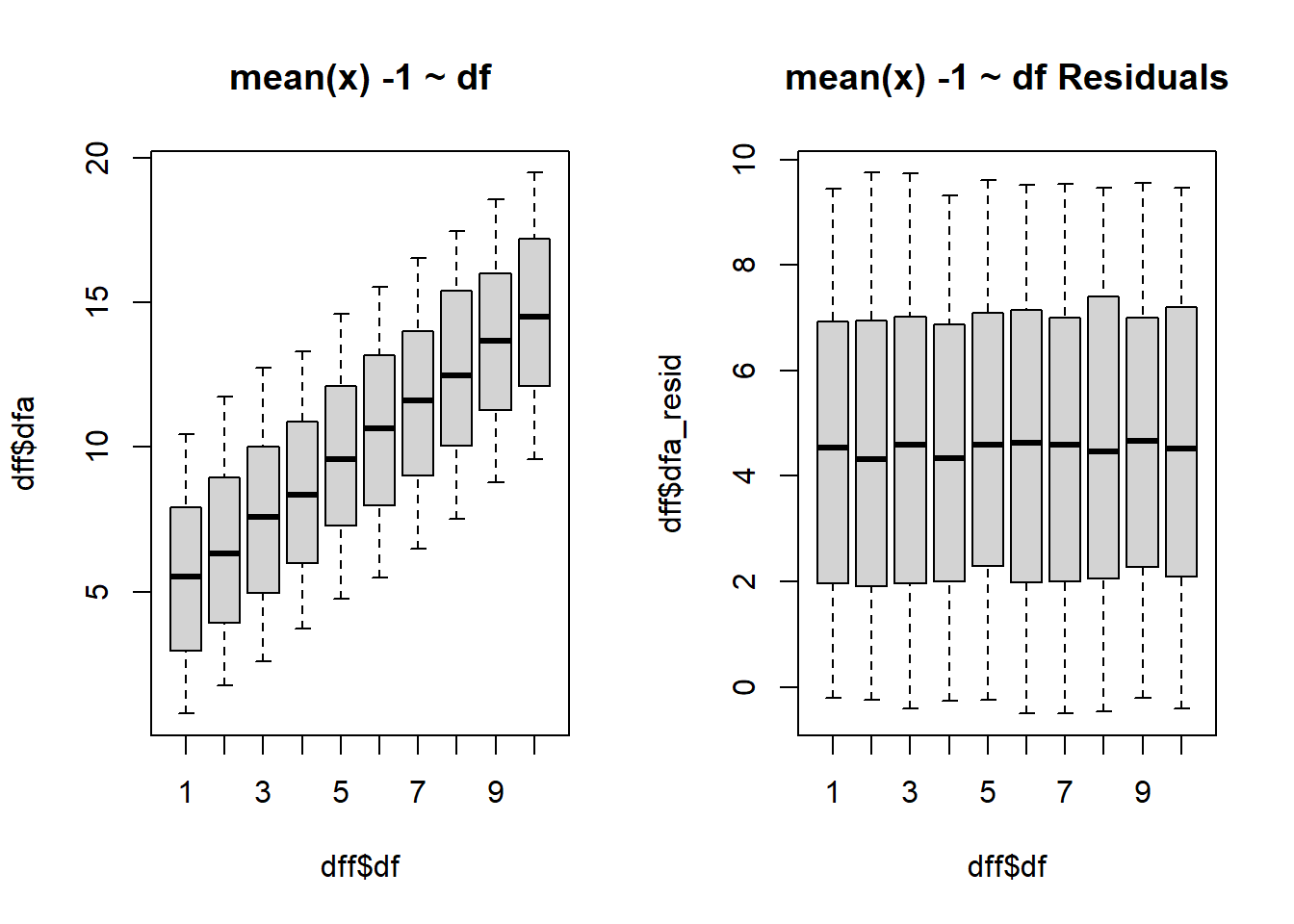
$ dfa <dbl> 0.9050589, 1.9826153, 3.0579375, 4.0515312, 4.2022289, 6.15…
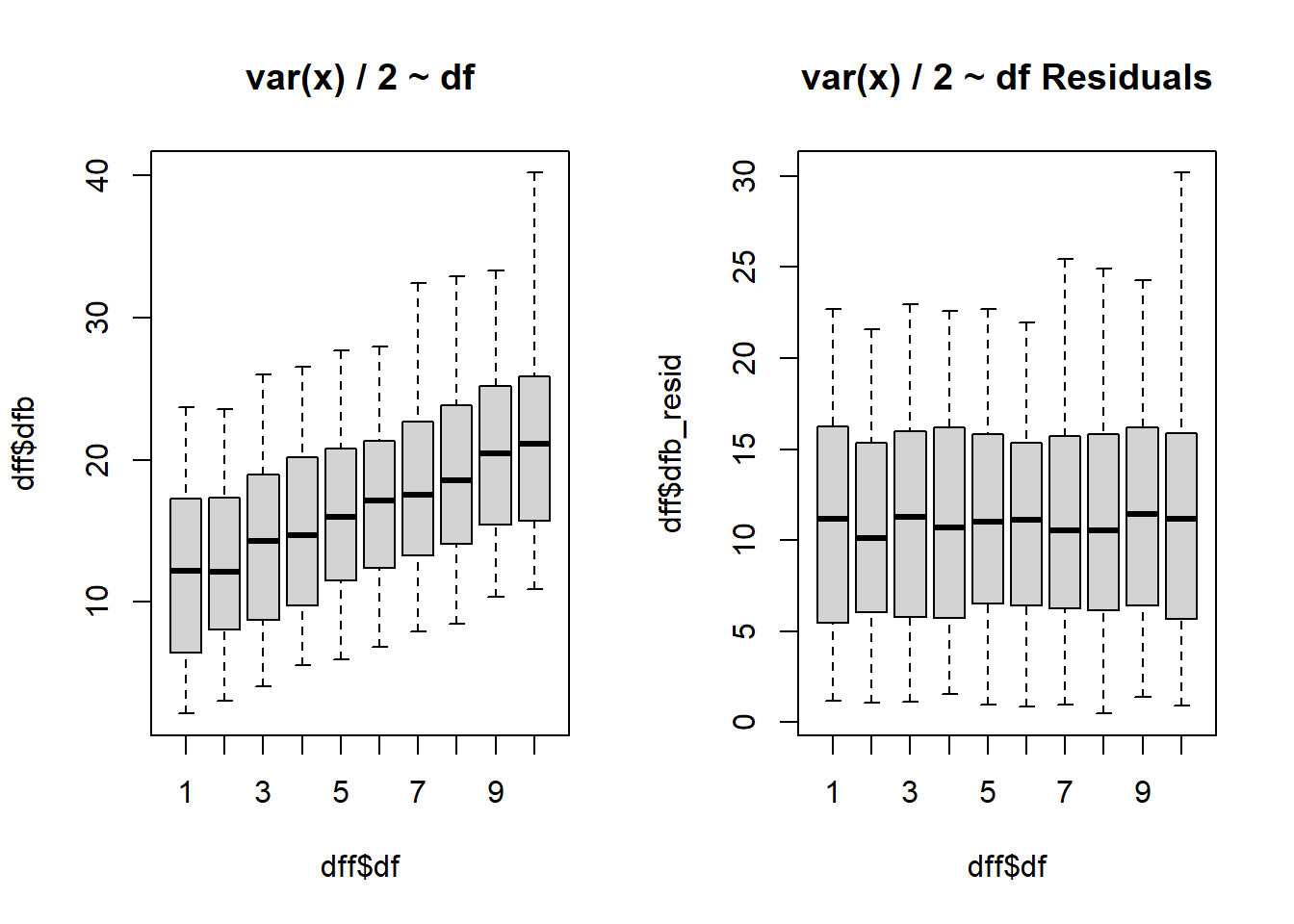
$ dfb <dbl> 2.626501, 5.428382, 7.297746, 9.265272, 8.465838, 14.597976…
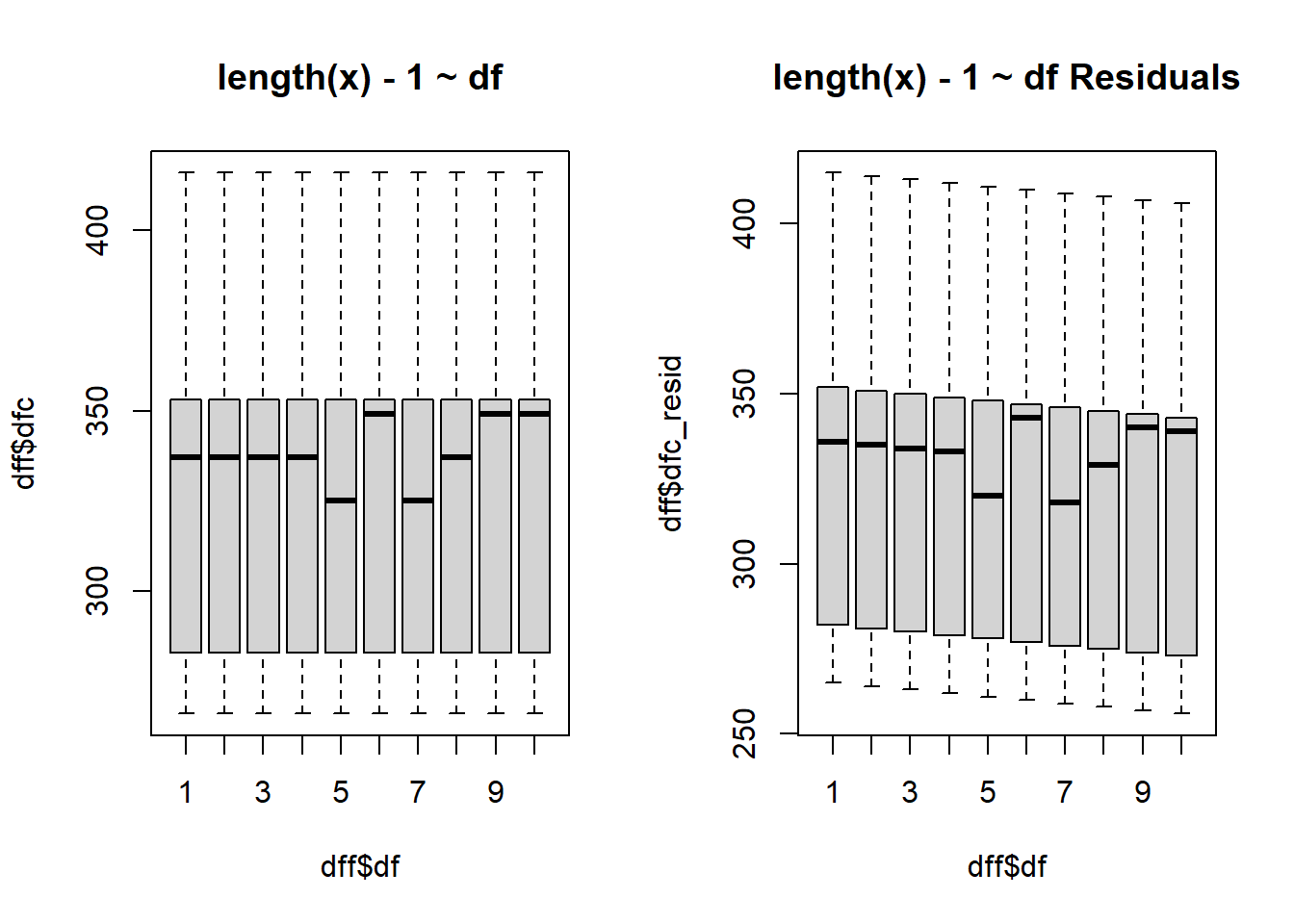
$ dfc <dbl> 283, 283, 283, 283, 283, 283, 283, 283, 283, 283, 283, 283,…
$ ncpa <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ ncpb <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
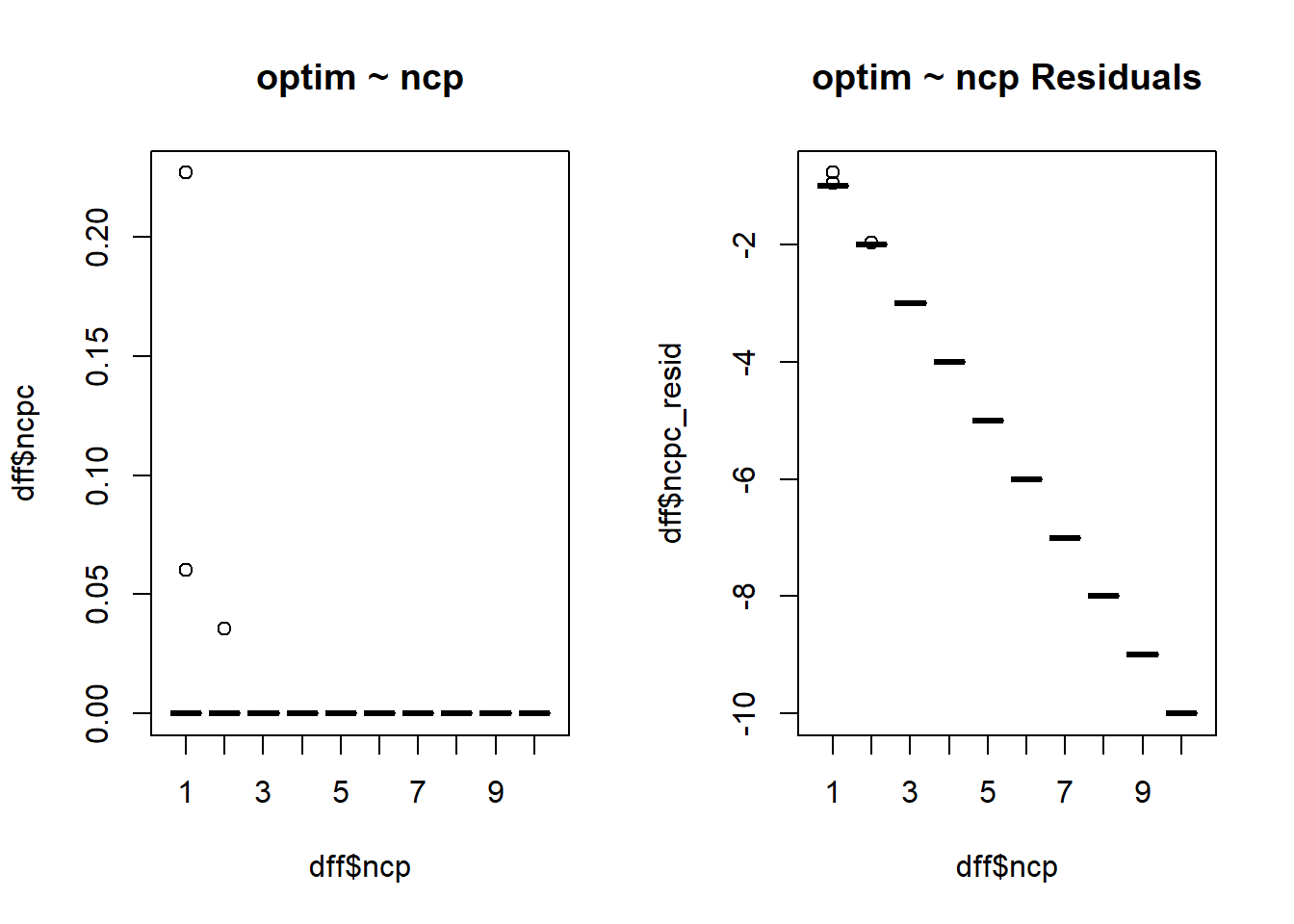
$ ncpc <dbl> 5.382789e-09, 8.170550e-09, 6.017177e-09, 8.618892e-09, 7.7…
$ dfa_resid <dbl> -0.09494109, 0.98261533, 2.05793748, 3.05153121, 3.20222890…
$ dfb_resid <dbl> 1.626501, 4.428382, 6.297746, 8.265272, 7.465838, 13.597976…
$ dfc_resid <dbl> 282, 282, 282, 282, 282, 282, 282, 282, 282, 282, 281, 281,…
$ dfd_resid <dbl> 0.177090434, -0.009400632, -0.020782073, -0.221812344, 0.51…
$ ncpa_resid <dbl> 0, -1, -2, -3, -4, -5, -6, -7, -8, -9, 0, -1, -2, -3, -4, -…
$ ncpb_resid <dbl> -1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -1, -2, -3, -4, -5…
$ ncpc_resid <dbl> -1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -1, -2, -3, -4, -5…
$ ncpd_resid <dbl> -0.27683618, -0.05376753, 0.03717560, 0.23474943, -1.238888…