Introduction
Many times in modeling we want to get the uncertainty in the model, well, bootstrapping to the rescue!
I am going to go over a very simple example on how to use purrr and modelr for this situation. We will use the mtcars dataset.
Examples
Let’s get right into it.
library(tidyverse)
library(tidymodels)
df <- mtcars
fit_boots <- df %>%
modelr::bootstrap(n = 200, id = 'boot_num') %>%
group_by(boot_num) %>%
mutate(fit = map(strap, ~lm(mpg ~ ., data = data.frame(.))))
fit_boots
# A tibble: 200 × 3
# Groups: boot_num [200]
strap boot_num fit
<list> <chr> <list>
1 <resample [32 x 11]> 001 <lm>
2 <resample [32 x 11]> 002 <lm>
3 <resample [32 x 11]> 003 <lm>
4 <resample [32 x 11]> 004 <lm>
5 <resample [32 x 11]> 005 <lm>
6 <resample [32 x 11]> 006 <lm>
7 <resample [32 x 11]> 007 <lm>
8 <resample [32 x 11]> 008 <lm>
9 <resample [32 x 11]> 009 <lm>
10 <resample [32 x 11]> 010 <lm>
# … with 190 more rows
Now lets get our parameter estimates.
# get parameters ####
params_boot <- fit_boots %>%
mutate(tidy_fit = map(fit, tidy)) %>%
unnest(cols = tidy_fit) %>%
ungroup()
# get predictions
preds_boot <- fit_boots %>%
mutate(augment_fit = map(fit, augment)) %>%
unnest(cols = augment_fit) %>%
ungroup()
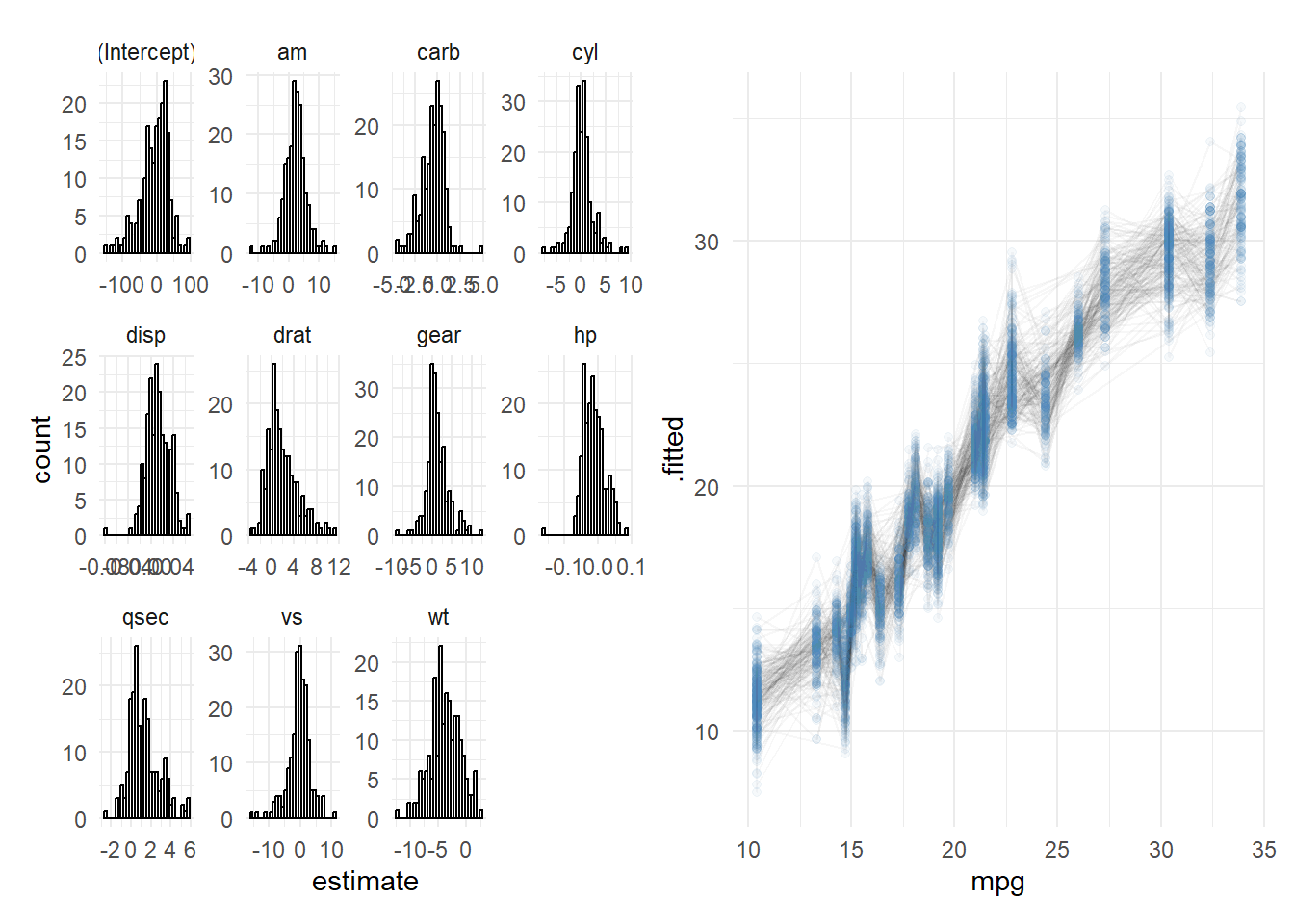
Time to visualize.
library(patchwork)
# plot distribution of estimated parameters
p1 <- ggplot(params_boot, aes(estimate)) +
geom_histogram(col = 'black', fill = 'white') +
facet_wrap(~ term, scales = 'free') +
theme_minimal()
# plot points with predictions
p2 <- ggplot() +
geom_line(aes(mpg, .fitted, group = boot_num), preds_boot, alpha = .03) +
geom_point(aes(mpg, .fitted), preds_boot, col = 'steelblue', alpha = 0.05) +
theme_minimal()
# plot both
p1 + p2
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Voila!