hai_knn_data_prepper(.data, .recipe_formula)Introduction
Minimal coding ML is not something that is unheard of and is rather prolific, think h2o and pycaret just to name two. There is also no shortage available for R with the h2o interface, and tidyfit. There are also similar low-code workflows in my r package {healthyR.ai}. Today I will specifically go through the workflow for Automatic KNN classification for the Iris data set where we will classify the Species.
Function
Let’s take a look at the two {healthyR.ai} functions that we will be using. First we have the data prepper hai_knn_data_prepper() which will get the data ready for use with the knn algorithm, and then we have the actual auto ml function hai_auto_knn(). Let’s take a look at the function calls.
First hai_knn_data_prepper()
Now let’s look at the arguments to those parameters.
.data- The data that you are passing to the function. Can be any type of data that is accepted by the data parameter of the recipes::reciep() function..recipe_formula- The formula that is going to be passed. For example if you are using the iris data then the formula would most likely be something likeSpecies ~ .
Now let’s take a look at the automl function.
hai_auto_knn(
.data,
.rec_obj,
.splits_obj = NULL,
.rsamp_obj = NULL,
.tune = TRUE,
.grid_size = 10,
.num_cores = 1,
.best_metric = "rmse",
.model_type = "regression"
)Again let’s look at the arguments to the parameters.
.data- The data being passed to the function. The time-series object..rec_obj- This is the recipe object you want to use. You can usehai_knn_data_prepper()an automatic recipe_object..splits_obj- NULL is the default, when NULL then one will be created..rsamp_obj- NULL is the default, when NULL then one will be created. It will default to creating an rsample::mc_cv() object..tune- Default is TRUE, this will create a tuning grid and tuned workflow.grid_size- Default is 10.num_cores- Default is 1.best_metric- Default is “rmse”. You can choose a metric depending on the model_type used. If regression then seehai_default_regression_metric_set(), if classification then seehai_default_classification_metric_set()..model_type- Default is regression, can also be classification.
Example
For this example we are going to use the iris data set where we are going to use the hai_auto_knn() to classify the Species.
library(healthyR.ai)
data <- iris
rec_obj <- hai_knn_data_prepper(data, Species ~ .)
auto_knn <- hai_auto_knn(
.data = data,
.rec_obj = rec_obj,
.best_metric = "f_meas",
.model_type = "classification",
.num_cores = 12
)Now let’s take a look at the complete output of the auto_knn object. The outputs are as follows:
- recipe
- model specification
- workflow
- tuned model (grid ect)
Recipe Output
auto_knn$recipe_infoRecipe
Inputs:
role #variables
outcome 1
predictor 4
Operations:
Novel factor level assignment for recipes::all_nominal_predictors()
Dummy variables from recipes::all_nominal_predictors()
Zero variance filter on recipes::all_predictors()
Centering and scaling for recipes::all_numeric()Model Info
auto_knn$model_info$was_tuned[1] "tuned"auto_knn$model_info$model_specK-Nearest Neighbor Model Specification (classification)
Main Arguments:
neighbors = tune::tune()
weight_func = tune::tune()
dist_power = tune::tune()
Computational engine: kknn auto_knn$model_info$wflw══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: nearest_neighbor()
── Preprocessor ────────────────────────────────────────────────────────────────
4 Recipe Steps
• step_novel()
• step_dummy()
• step_zv()
• step_normalize()
── Model ───────────────────────────────────────────────────────────────────────
K-Nearest Neighbor Model Specification (classification)
Main Arguments:
neighbors = tune::tune()
weight_func = tune::tune()
dist_power = tune::tune()
Computational engine: kknn auto_knn$model_info$fitted_wflw══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: nearest_neighbor()
── Preprocessor ────────────────────────────────────────────────────────────────
4 Recipe Steps
• step_novel()
• step_dummy()
• step_zv()
• step_normalize()
── Model ───────────────────────────────────────────────────────────────────────
Call:
kknn::train.kknn(formula = ..y ~ ., data = data, ks = min_rows(13L, data, 5), distance = ~1.69935879141092, kernel = ~"rank")
Type of response variable: nominal
Minimal misclassification: 0.03571429
Best kernel: rank
Best k: 13Tuning Info
auto_knn$tuned_info$tuning_grid# A tibble: 10 × 3
neighbors weight_func dist_power
<int> <chr> <dbl>
1 4 triangular 0.764
2 11 rectangular 0.219
3 5 gaussian 1.35
4 14 triweight 0.351
5 5 biweight 1.05
6 9 optimal 1.87
7 7 cos 0.665
8 11 inv 1.18
9 13 rank 1.70
10 1 epanechnikov 1.58 auto_knn$tuned_info$cv_obj# Monte Carlo cross-validation (0.75/0.25) with 25 resamples
# A tibble: 25 × 2
splits id
<list> <chr>
1 <split [84/28]> Resample01
2 <split [84/28]> Resample02
3 <split [84/28]> Resample03
4 <split [84/28]> Resample04
5 <split [84/28]> Resample05
6 <split [84/28]> Resample06
7 <split [84/28]> Resample07
8 <split [84/28]> Resample08
9 <split [84/28]> Resample09
10 <split [84/28]> Resample10
# … with 15 more rowsauto_knn$tuned_info$tuned_results# Tuning results
# Monte Carlo cross-validation (0.75/0.25) with 25 resamples
# A tibble: 25 × 4
splits id .metrics .notes
<list> <chr> <list> <list>
1 <split [84/28]> Resample01 <tibble [110 × 7]> <tibble [0 × 3]>
2 <split [84/28]> Resample02 <tibble [110 × 7]> <tibble [0 × 3]>
3 <split [84/28]> Resample03 <tibble [110 × 7]> <tibble [0 × 3]>
4 <split [84/28]> Resample04 <tibble [110 × 7]> <tibble [0 × 3]>
5 <split [84/28]> Resample05 <tibble [110 × 7]> <tibble [0 × 3]>
6 <split [84/28]> Resample06 <tibble [110 × 7]> <tibble [0 × 3]>
7 <split [84/28]> Resample07 <tibble [110 × 7]> <tibble [0 × 3]>
8 <split [84/28]> Resample08 <tibble [110 × 7]> <tibble [0 × 3]>
9 <split [84/28]> Resample09 <tibble [110 × 7]> <tibble [0 × 3]>
10 <split [84/28]> Resample10 <tibble [110 × 7]> <tibble [0 × 3]>
# … with 15 more rowsauto_knn$tuned_info$grid_size[1] 10auto_knn$tuned_info$best_metric[1] "f_meas"auto_knn$tuned_info$best_result_set# A tibble: 1 × 9
neighbors weight_func dist_power .metric .estima…¹ mean n std_err .config
<int> <chr> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 13 rank 1.70 f_meas macro 0.957 25 0.00655 Prepro…
# … with abbreviated variable name ¹.estimatorauto_knn$tuned_info$tuning_grid_plot`geom_smooth()` using method = 'loess' and formula = 'y ~ x'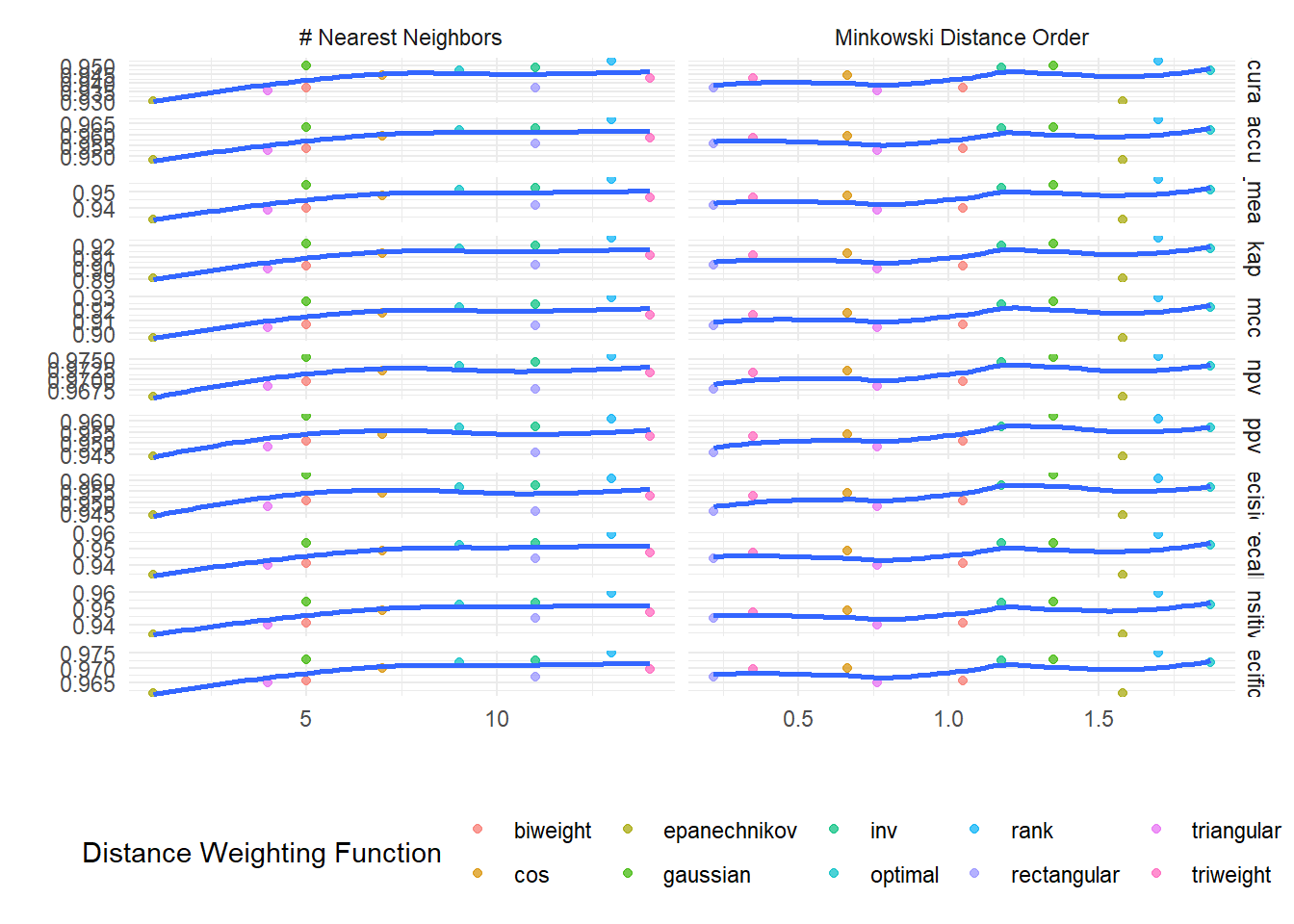
auto_knn$tuned_info$plotly_grid_plotVoila!
Thank you for reading.