x <- mtcars$mpg
x [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4
[16] 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7
[31] 15.0 21.4Steven P. Sanderson II, MPH
November 4, 2022
Many times in the real world we have a data set which is actually a sample as we typically do not know what the actual population is. This is where bootstrapping tends to come into play. It allows us to get a hold on what the possible parameter values are by taking repeated samples of the data that is available to us.
At it’s core it is a resampling method with replacement where it assigns measures of accuracy to the sample estimates. Here is the Wikipedia Article for bootstrapping.
In this post I am going to go over how to use the bootstrap function set with {TidyDensity}. You can find the pkgdown site with all function references here: TidyDensity
The first thing we will need is a dataset, and for this we are going to pick on the mtcars dataset and more specifically the mpg column. So let’s get to it!
[1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4
[16] 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7
[31] 15.0 21.4We see that x is a numeric vector, which is what we need for these {TidyDensity} functions. The first function that x will be put through is a function called tidy_bootstrap() Let’s take a look at the full function call and parameters of this function.
What you see above are the defaults for the function. Now lets go through the parameters.
.x - This is of course the numeric vector that you are passing to the function, in our case right now, it is x which we set to mtcars$mpg
.num_sims - This is how many simulations you want to run of x. This is done with replacement. So this is dictating how many bootstrap samples of x we want to take.
.proportion - How much of the data do you want to sample? The default here is 80%
.distribution_type - What kind of distribution are you sampling from? Is it a continuous or discrete distribution. This is important for plotting.
The function returns a tibble with the bootstrap column as a list object. Lets take a look at tidy_bootstrap(x). We are going to set simulations to 50 instead of the default 2000.
# A tibble: 50 × 2
sim_number bootstrap_samples
<fct> <list>
1 1 <dbl [25]>
2 2 <dbl [25]>
3 3 <dbl [25]>
4 4 <dbl [25]>
5 5 <dbl [25]>
6 6 <dbl [25]>
7 7 <dbl [25]>
8 8 <dbl [25]>
9 9 <dbl [25]>
10 10 <dbl [25]>
# … with 40 more rowsThe column bootstrap_samples holds the bootstrapped resamples of x at the given .proportion, in this instance, 80%.
From this point we can go straight into use the bootstrap_stat_plot() function if we choose. Under-the-hood it will make use of bootstrap_unnest_tbl(). All this function does is act as a helper to unnest the bootstrap_samples column of the returned tibble from tidy_bootstrap() Let’s take a look below.
# A tibble: 1,250 × 2
sim_number y
<fct> <dbl>
1 1 22.8
2 1 15.5
3 1 21.5
4 1 15
5 1 10.4
6 1 22.8
7 1 16.4
8 1 30.4
9 1 26
10 1 19.2
# … with 1,240 more rowsNow let’s get into the bootstrap_stat_plot() function of {TidyDensity}
The function bootstrap_stat_plot() was designed to handle data either from the tidy_bootstrap() or bootstrap_unnest_tbl() functions only. This was to ensure that the right type of data was being passed in and to ensure that the right type of output was guaranteed.
Let’s take a full look at the function call.
There are a few interesting parameters here, but like before we will go through all of them.
.data - This is the data that gets passed from either tidy_bootstrap() or bootstrap_unnest_tbl()
.value - This is the column from bootstrap_unnest_tbl() that you want to visualize, this is typically y
.stat - There are multiple cumulative stats that will work with this plot. These are all built directly into the {TidyDensity} package. You can find the supported ones that are built into this package at the reference page.
.show_groups - Do you want to show all of the simulation groups TRUE/FALSE
.show_ci_labels - If set to TRUE then the confidence interval labels will be shows on the graph as the final value.
.interactive - Do you want a plotly plot? Who doesn’t?
Now let’s walk though a few examples.
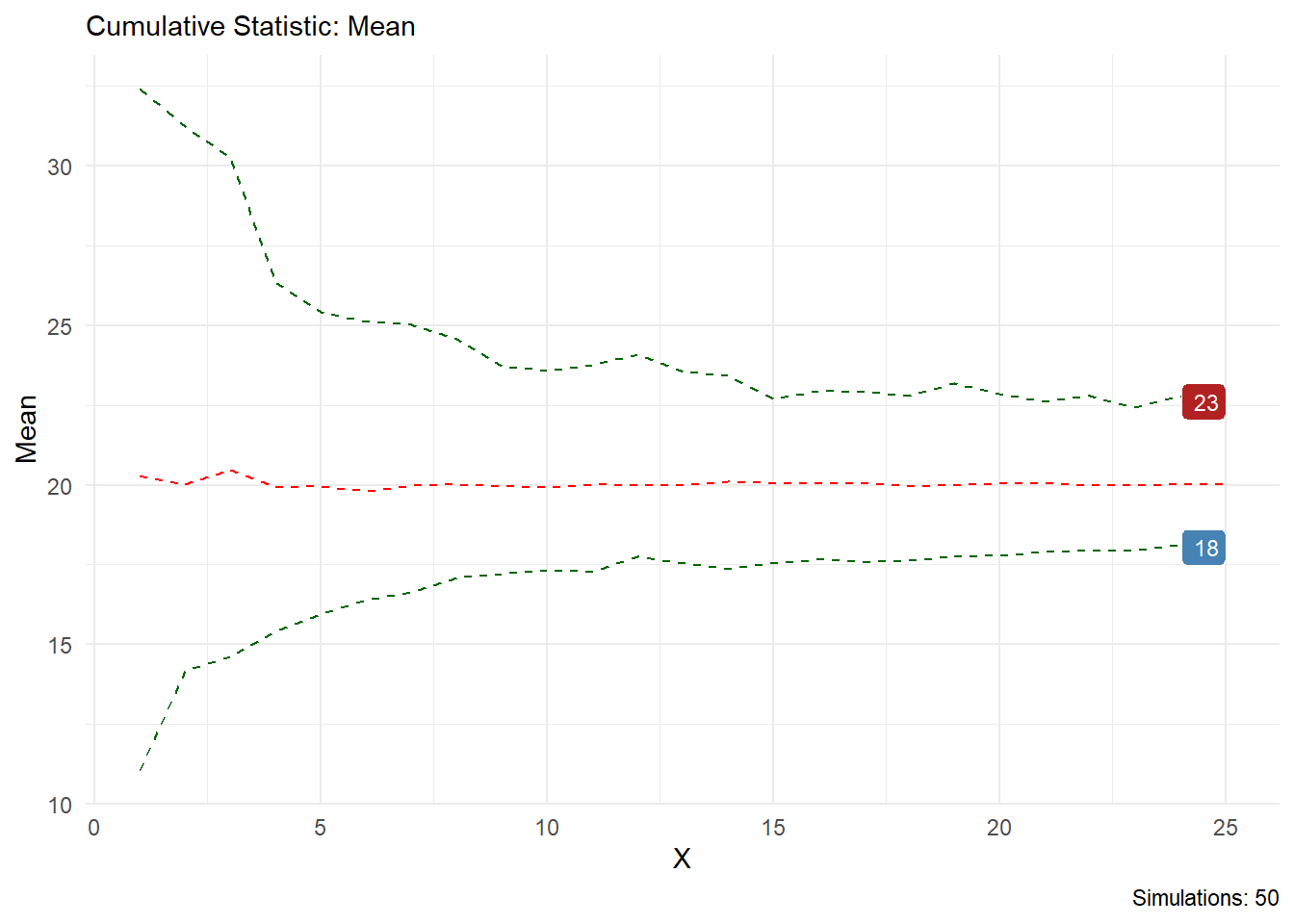
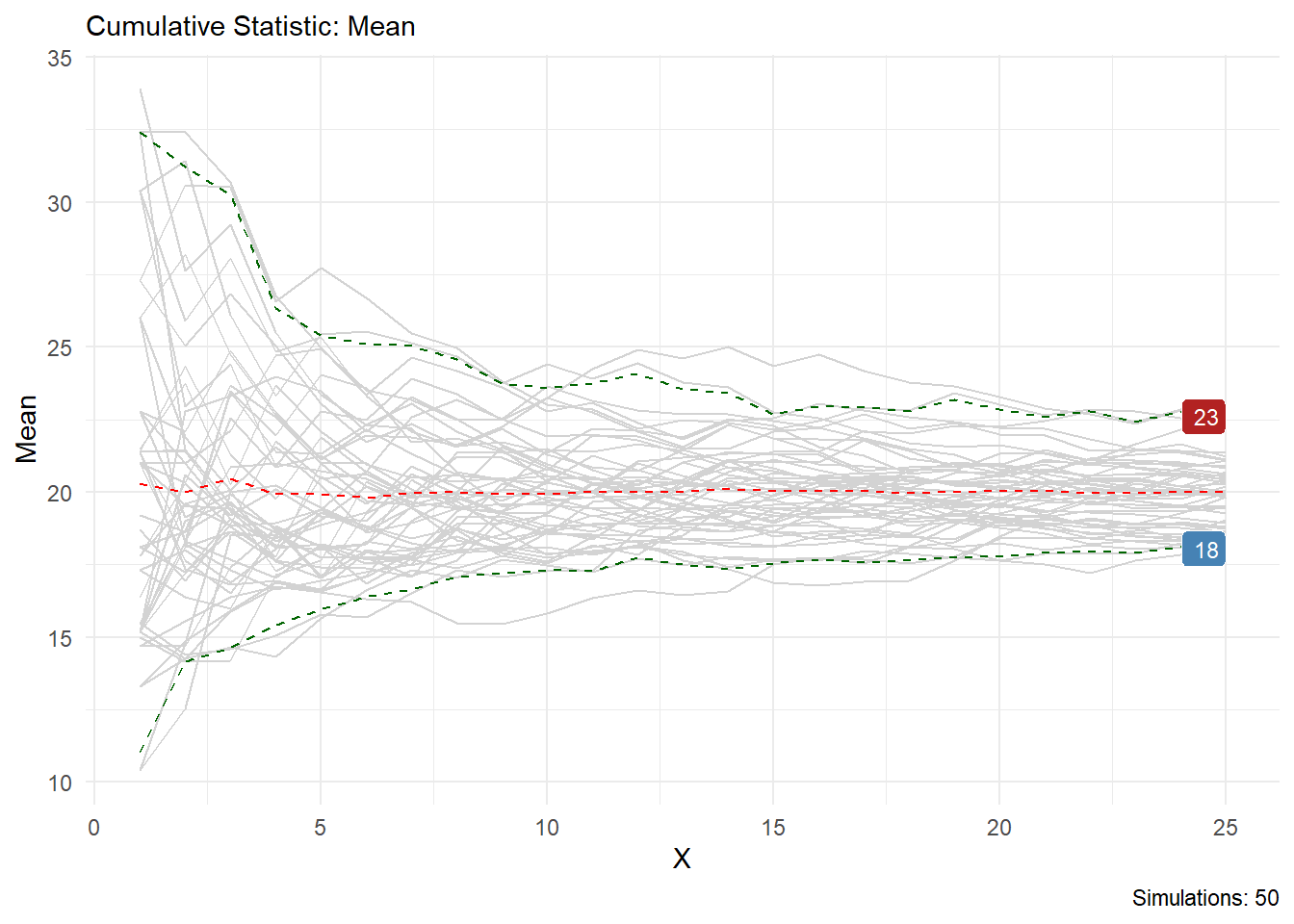
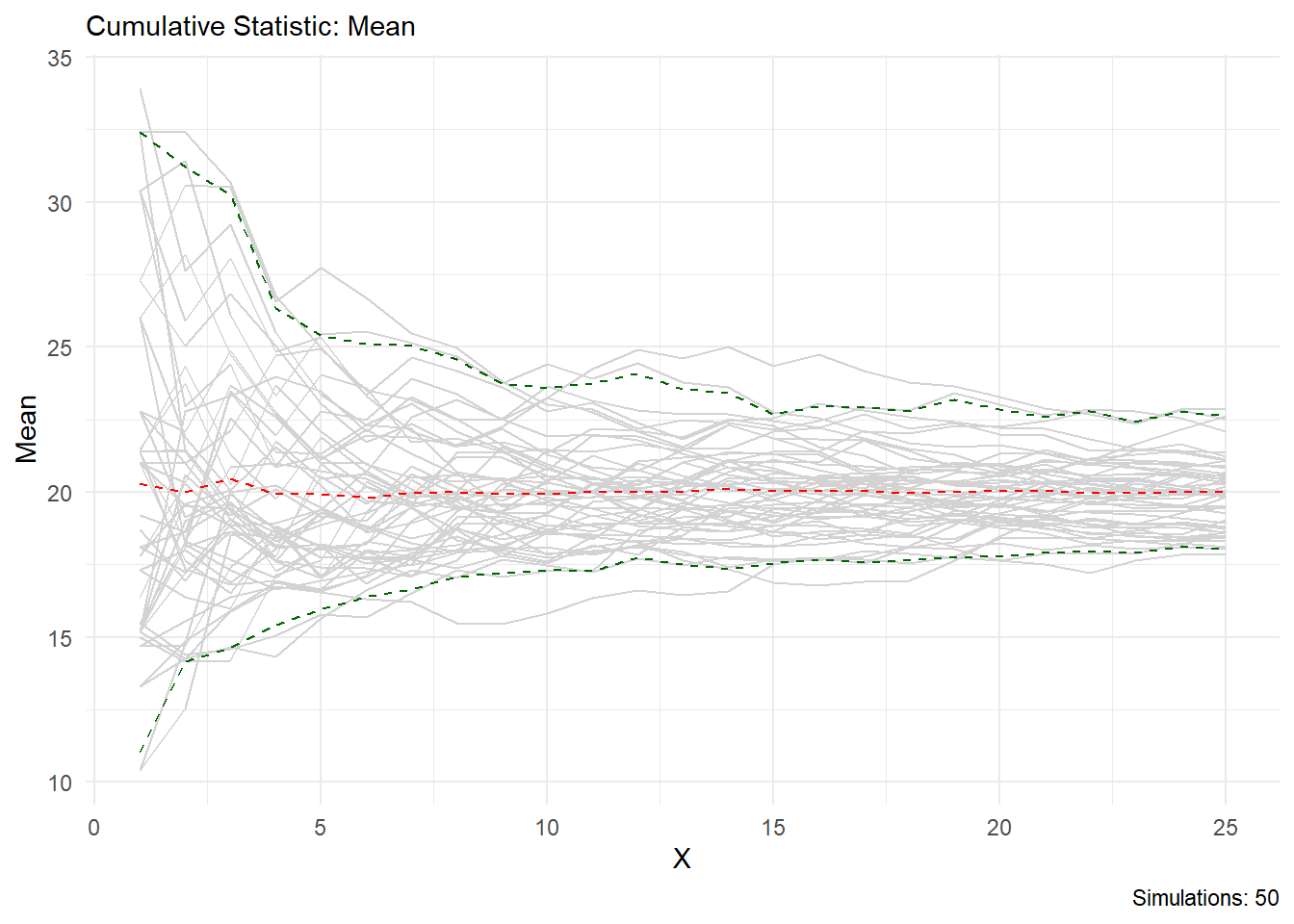
You can see from this output that the statistic you choose is printed in the chart title and on the y axis, the caption will also tell you how many simulations are present. Lets look at skewness as another example.
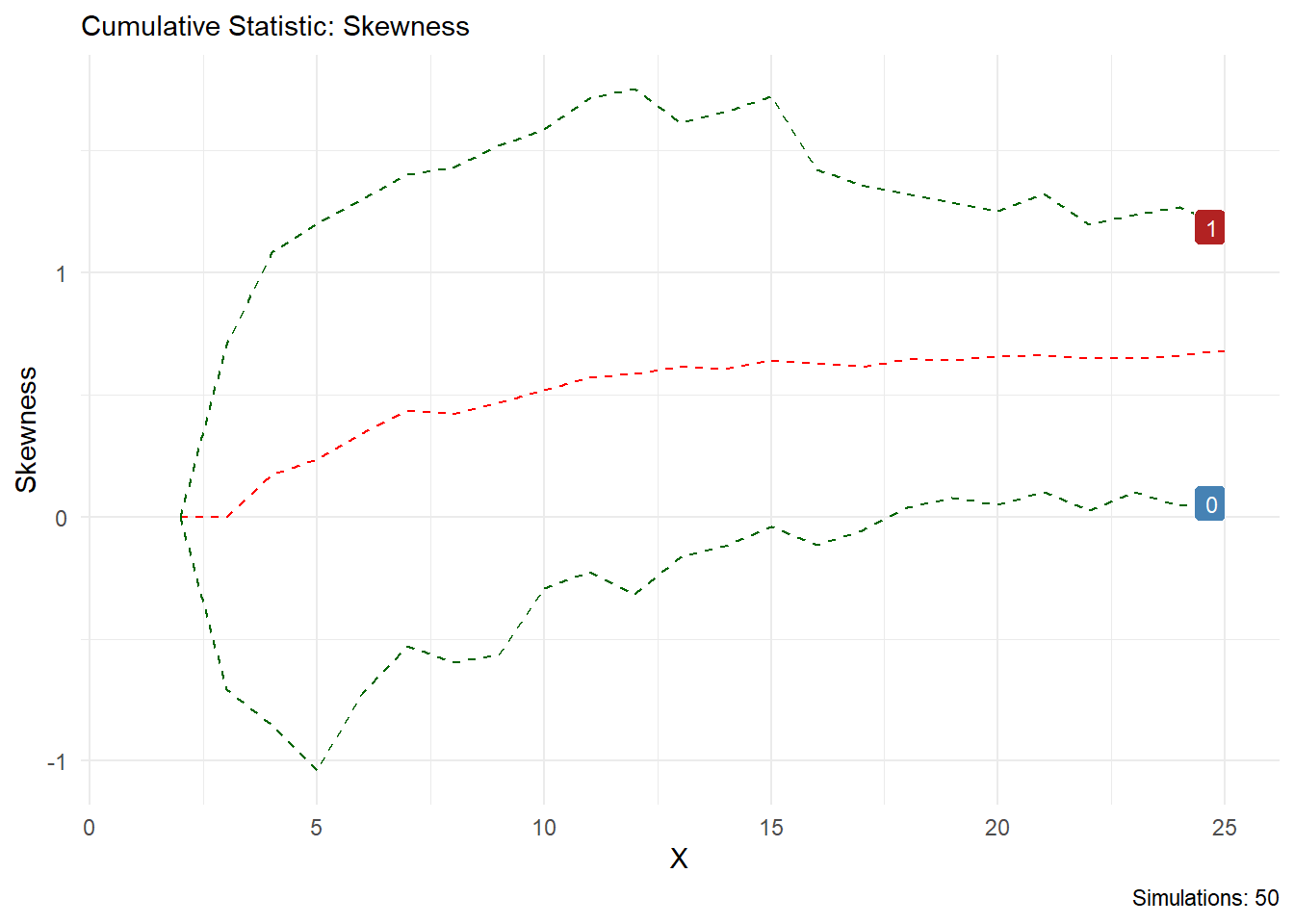
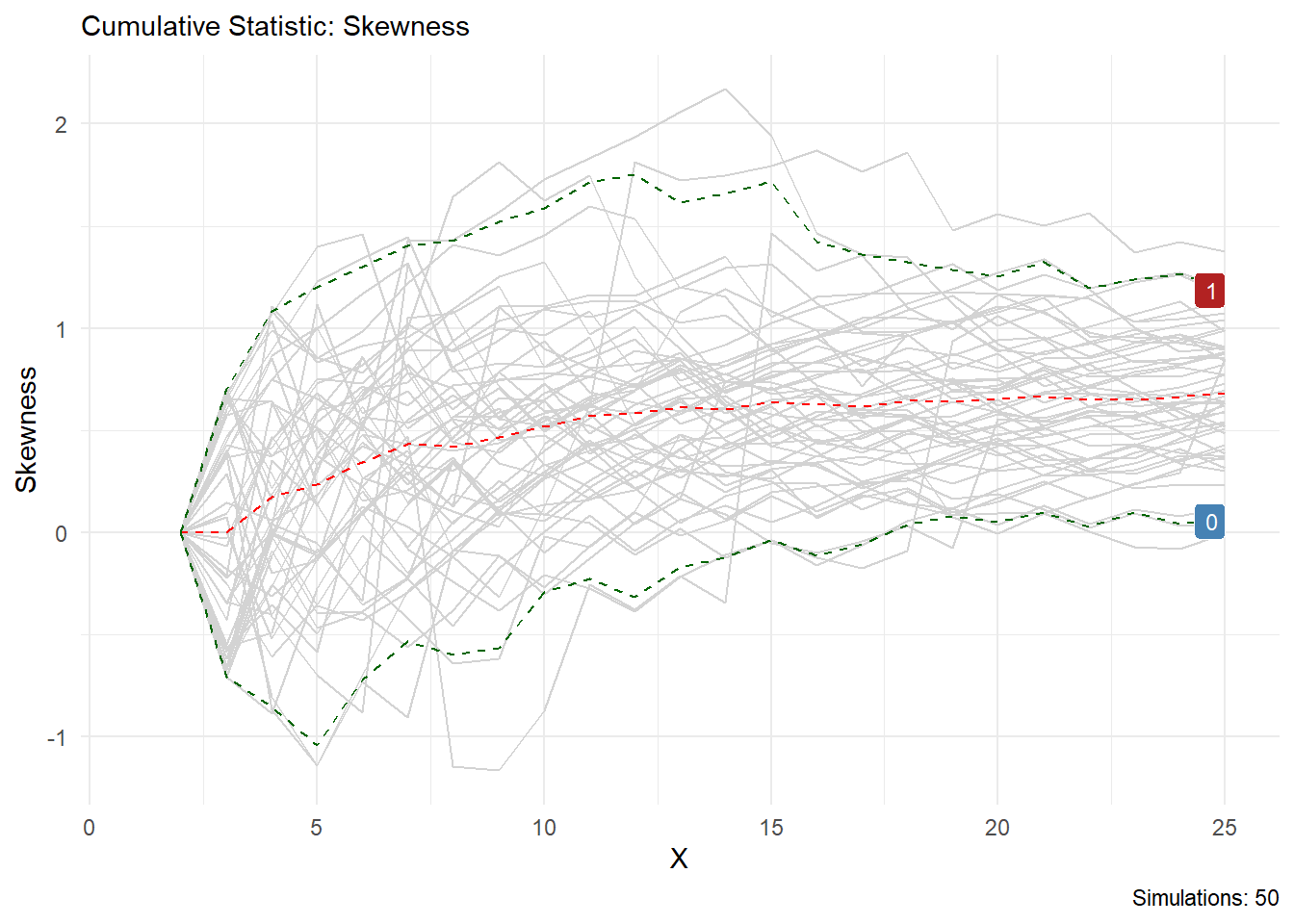
Volia!