ts_lag_correlation(
.data,
.date_col,
.value_col,
.lags = 1,
.heatmap_color_low = "white",
.heatmap_color_hi = "steelblue"
)Introduction
In time series analysis there is something called a lag. This simply means we take a look at some past event from some point in time t. This is a non-statistical method for looking at a relationship between a timeseries and its lags.
{healthyR.ts} has a function called ts_lag_correlation(). This function, as described by it’s name, provides more than just a simple lag plot.
This function provides a lot of extra information for the end user. First let’s go over the function call.
Function
Function Call
Here is the full call:
Here are the arguments that get supplied to the different parameters.
.data- A tibble of time series data.date_col- A date column.value_col- The value column being analyzed.lags- This is a vector of integer lags, ie 1 or c(1,6,12).heatmap_color_low- What color should the low values of the heatmap of the correlation matrix be, the default is ‘white’.heatmap_color_hi- What color should the low values of the heatmap of the correlation matrix be, the default is ‘steelblue’
Function Return
The function itself returns a list object. The list has the following elements in it:
Data Elements
lag_listlag_tblcorrelation_lag_matrixcorrelation_lag_tbl
Plot Elements
lag_plotplotly_lag_plotcorrelation_heatmapplotly_heatmap
Example
Let’s take a look at an example. We are going to use the built in data set of AirPassengers.
library(healthyR.ts)
library(dplyr)
df <- ts_to_tbl(AirPassengers) %>% select(-index)
lags <- c(1,3,6,12)
output <- ts_lag_correlation(
.data = df,
.date_col = date_col,
.value_col = value,
.lags = lags
)Now that we have our output, let’s examine each piece of it.
Data Elements
Here are the data elements.
output$data$lag_list[[1]]
# A tibble: 143 × 3
lag value lagged_value
<fct> <dbl> <dbl>
1 1 118 112
2 1 132 118
3 1 129 132
4 1 121 129
5 1 135 121
6 1 148 135
7 1 148 148
8 1 136 148
9 1 119 136
10 1 104 119
# … with 133 more rows
[[2]]
# A tibble: 141 × 3
lag value lagged_value
<fct> <dbl> <dbl>
1 3 129 112
2 3 121 118
3 3 135 132
4 3 148 129
5 3 148 121
6 3 136 135
7 3 119 148
8 3 104 148
9 3 118 136
10 3 115 119
# … with 131 more rows
[[3]]
# A tibble: 138 × 3
lag value lagged_value
<fct> <dbl> <dbl>
1 6 148 112
2 6 148 118
3 6 136 132
4 6 119 129
5 6 104 121
6 6 118 135
7 6 115 148
8 6 126 148
9 6 141 136
10 6 135 119
# … with 128 more rows
[[4]]
# A tibble: 132 × 3
lag value lagged_value
<fct> <dbl> <dbl>
1 12 115 112
2 12 126 118
3 12 141 132
4 12 135 129
5 12 125 121
6 12 149 135
7 12 170 148
8 12 170 148
9 12 158 136
10 12 133 119
# … with 122 more rowsThis is a list of all the tibbles of the different lags that were chosen.
output$data$lag_tbl# A tibble: 554 × 4
lag value lagged_value lag_title
<fct> <dbl> <dbl> <fct>
1 1 118 112 Lag: 1
2 1 132 118 Lag: 1
3 1 129 132 Lag: 1
4 1 121 129 Lag: 1
5 1 135 121 Lag: 1
6 1 148 135 Lag: 1
7 1 148 148 Lag: 1
8 1 136 148 Lag: 1
9 1 119 136 Lag: 1
10 1 104 119 Lag: 1
# … with 544 more rowsThis is the long lag tibble with all of the lags in it.
output$data$correlation_lag_matrix value value_lag1 value_lag3 value_lag6 value_lag12
value 1.0000000 0.9542938 0.8186636 0.7657001 0.9905274
value_lag1 0.9542938 1.0000000 0.8828054 0.7726530 0.9492382
value_lag3 0.8186636 0.8828054 1.0000000 0.8349550 0.8218493
value_lag6 0.7657001 0.7726530 0.8349550 1.0000000 0.7780911
value_lag12 0.9905274 0.9492382 0.8218493 0.7780911 1.0000000This is the correlation matrix.
output$data$correlation_lag_tbl# A tibble: 25 × 3
name data_names value
<fct> <fct> <dbl>
1 value value 1
2 value_lag1 value 0.954
3 value_lag3 value 0.819
4 value_lag6 value 0.766
5 value_lag12 value 0.991
6 value value_lag1 0.954
7 value_lag1 value_lag1 1
8 value_lag3 value_lag1 0.883
9 value_lag6 value_lag1 0.773
10 value_lag12 value_lag1 0.949
# … with 15 more rowsThis is the correlation lag tibble
Plot Elements
output$plots$lag_plot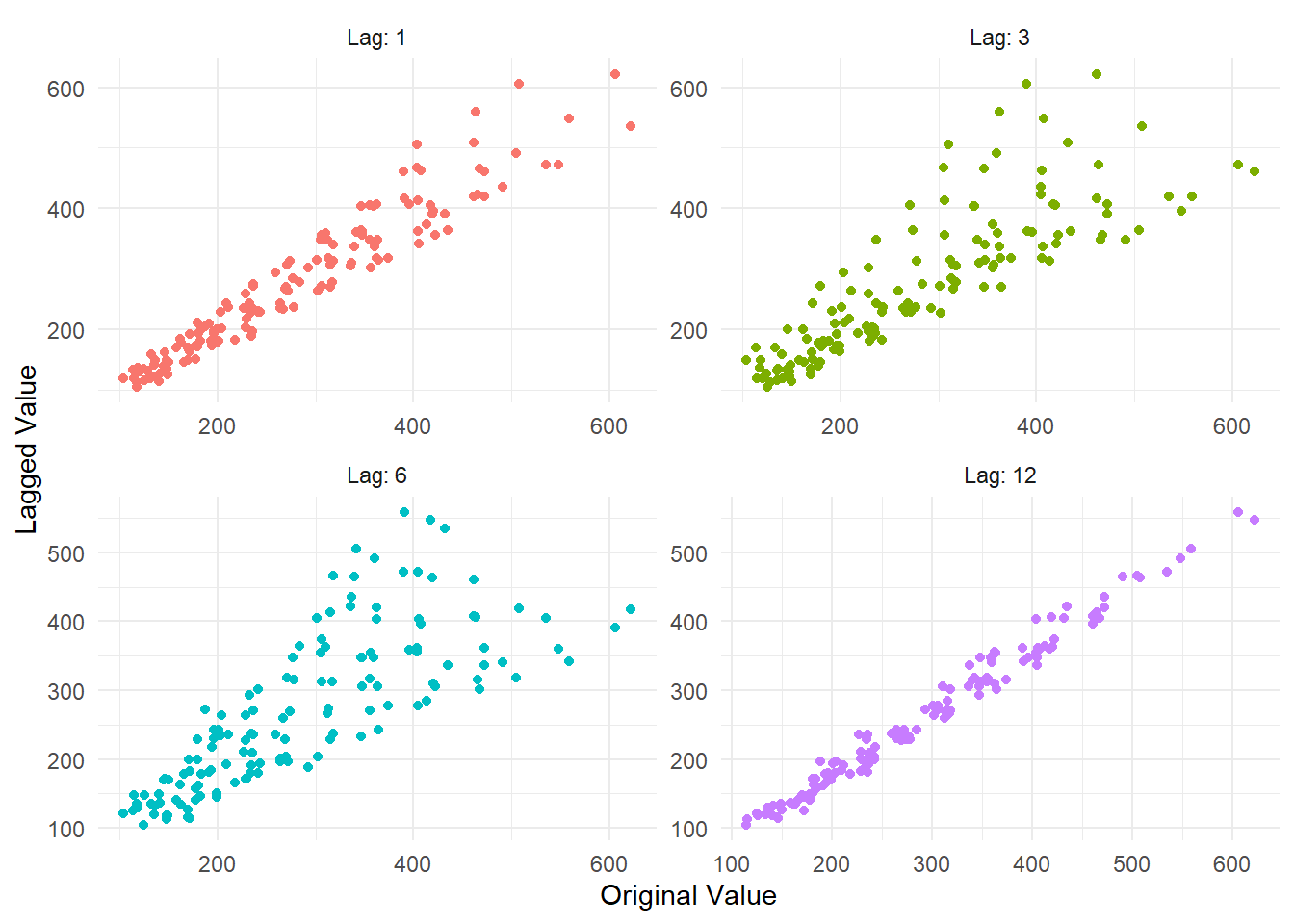
The Lag Plot itself.
output$plots$plotly_lag_plotA plotly version of the lag plot.
output$plots$correlation_heatmap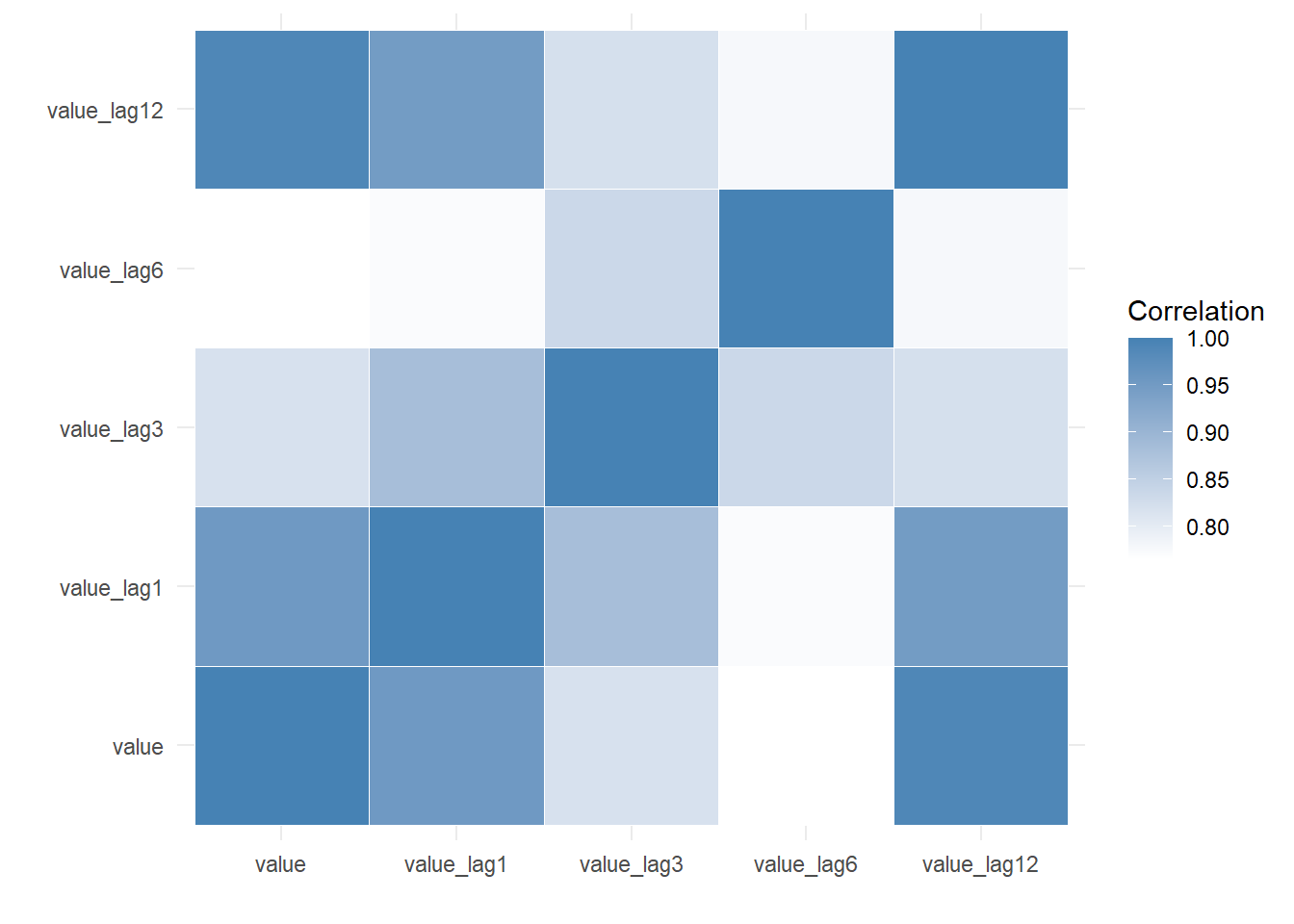
A heatmap of the correlations.
output$plots$plotly_heatmapA plotly version of the correlation heatmap.
Voila!