Using Tidy FFT
Analyze Time Series data with tidy_fft
Steven P. Sanderson II, MPH
2025-12-09
Source:vignettes/using-tidy-fft.Rmd
using-tidy-fft.Rmd
library(healthyR.ts)
#>
#> == Welcome to healthyR.ts ===========================================================================
#> If you find this package useful, please leave a star:
#> https://github.com/spsanderson/healthyR.ts'
#>
#> If you encounter a bug or want to request an enhancement please file an issue at:
#> https://github.com/spsanderson/healthyR.ts/issues
#>
#> Thank you for using healthyR.tsIntroduction
In this vignette we will discuss how to use the tidy_fft
function, what it does, and what it produces.
The Function
The tidy_fft function has only a few parameters, six to
be exact. There are some sensible defaults made. It is important that
when you use this function, that you supply it with a full time-series
data set, one that has no missing data in it as this will affect your
results.
Funcation and Parameters
The function and its full parameters are as follows:
tidy_fft(
.data,
.date_col,
.value_col,
.frequency = 12L,
.harmonics = 1L,
.upsampling = 10L
)The .data argument is the actual formatted data that
will get passed to the function, the time series data. The
.date_col argument is the column that holds the datetime of
interest. The .value column is the column that holds the
value that is being analyzed by the function, this can be counts,
averages, any type of value that is in the time series. The
.frequency argument details the cyclical nature of the
data, is it 12 for monthly, 7 for weekly, etc. The
.harmonics argument will tell the function how many times
the fft should be run internally and how many filters
should be made. Finally the .upsampling argument will tell
the function how much the function should up sample the time
parameter.
Let us now work through a simple example.
Example
Data
Lets get started with some data.
suppressPackageStartupMessages(library(dplyr))
suppressPackageStartupMessages(library(ggplot2))
suppressPackageStartupMessages(library(timetk))
data_tbl <- AirPassengers %>%
ts_to_tbl() %>%
select(-index)Now that we have our sample data, let’s check it out.
glimpse(data_tbl)
#> Rows: 144
#> Columns: 2
#> $ date_col <date> 1949-01-01, 1949-02-01, 1949-03-01, 1949-04-01, 1949-05-01, …
#> $ value <dbl> 112, 118, 132, 129, 121, 135, 148, 148, 136, 119, 104, 118, 1…Plot Data
Lets take a look at a time series plot of the data.
suppressPackageStartupMessages(library(timetk))
data_tbl %>%
plot_time_series(
.date_var = date_col,
.value = value
)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the timetk package.
#> Please report the issue at
#> <https://github.com/business-science/timetk/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Ignoring unknown labels:
#> • colour : "Legend"Now that we know what our data looks like, lets go ahead and run it
through the function and assign it to a variable called
output
Run Function
output <- tidy_fft(
.data = data_tbl,
.date_col = date_col,
.value_col = value,
.harmonics = 8,
.frequency = 12,
.upsampling = 5
)Now that we have run the function, let’s take a look at the output.
Output
The function invisibly returns a list object, hence the need to assign it to a variable. There are a total of 4 different sections of data in the list that are returned. These are:
- data
- plots
- parameters
- model
Output Data
In this section we will go over all of the data components that are
returned. We can access all of the data in the usual format
output$data, which in of itself will return another list of
objects, 7 to be specific. Lets go through them all.
data
The data element accessed by output$data$data is the
original data with a few elements added to it. Let’s take a look:
output$data$data %>%
glimpse()
#> Rows: 5,760
#> Columns: 6
#> $ harmonic <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ time <dbl> 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.2, 2.4, 2.6, 2.8, 3.0, 3.2,…
#> $ y_actual <dbl> 112, NA, NA, NA, NA, 118, NA, NA, NA, NA, 132, NA, NA, NA, …
#> $ y_hat <dbl> 292.1741, 291.0941, 290.0134, 288.9318, 287.8497, 286.7669,…
#> $ x <dbl> 1, 0, 0, 0, 0, 2, 0, 0, 0, 0, 3, 0, 0, 0, 0, 4, 0, 0, 0, 0,…
#> $ error_term <dbl> -180.1741, NA, NA, NA, NA, -168.7669, NA, NA, NA, NA, -149.…error_data
The error_data element accessed by
output$data$error_data is a tibble that has
the original data, plus a few other elements and an error term that is
the actual value minus the harmonic output. This is done for each
harmonic level.
output$data$error_data %>%
glimpse()
#> Rows: 1,152
#> Columns: 6
#> $ harmonic <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ time <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, …
#> $ y_actual <dbl> 112, 118, 132, 129, 121, 135, 148, 148, 136, 119, 104, 118,…
#> $ y_hat <dbl> 292.1741, 286.7669, 281.3475, 275.9261, 270.5130, 265.1185,…
#> $ x <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, …
#> $ error_term <dbl> -180.17406, -168.76694, -149.34751, -146.92608, -149.51298,…input_vector
The input_vector is just the value column that was passed to the function.
output$data$input_vector
#> [1] 112 118 132 129 121 135 148 148 136 119 104 118 115 126 141 135 125 149
#> [19] 170 170 158 133 114 140 145 150 178 163 172 178 199 199 184 162 146 166
#> [37] 171 180 193 181 183 218 230 242 209 191 172 194 196 196 236 235 229 243
#> [55] 264 272 237 211 180 201 204 188 235 227 234 264 302 293 259 229 203 229
#> [73] 242 233 267 269 270 315 364 347 312 274 237 278 284 277 317 313 318 374
#> [91] 413 405 355 306 271 306 315 301 356 348 355 422 465 467 404 347 305 336
#> [109] 340 318 362 348 363 435 491 505 404 359 310 337 360 342 406 396 420 472
#> [127] 548 559 463 407 362 405 417 391 419 461 472 535 622 606 508 461 390 432maximum_harmonic_tbl
The maximum_harmonic_tbl is a tibble that has data
regarding the maximum harmonic entered into the function, this will be
the most flexible data returned.
output$data$maximum_harmonic_tbl %>%
glimpse()
#> Rows: 720
#> Columns: 6
#> $ harmonic <fct> 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8,…
#> $ time <dbl> 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.2, 2.4, 2.6, 2.8, 3.0, 3.2,…
#> $ y_actual <dbl> 112, NA, NA, NA, NA, 118, NA, NA, NA, NA, 132, NA, NA, NA, …
#> $ y_hat <dbl> 288.7745, 279.8566, 270.9787, 262.1584, 253.4132, 244.7606,…
#> $ x <dbl> 1, 0, 0, 0, 0, 2, 0, 0, 0, 0, 3, 0, 0, 0, 0, 4, 0, 0, 0, 0,…
#> $ error_term <dbl> -176.77449, NA, NA, NA, NA, -126.76057, NA, NA, NA, NA, -71…differenced_value_tbl
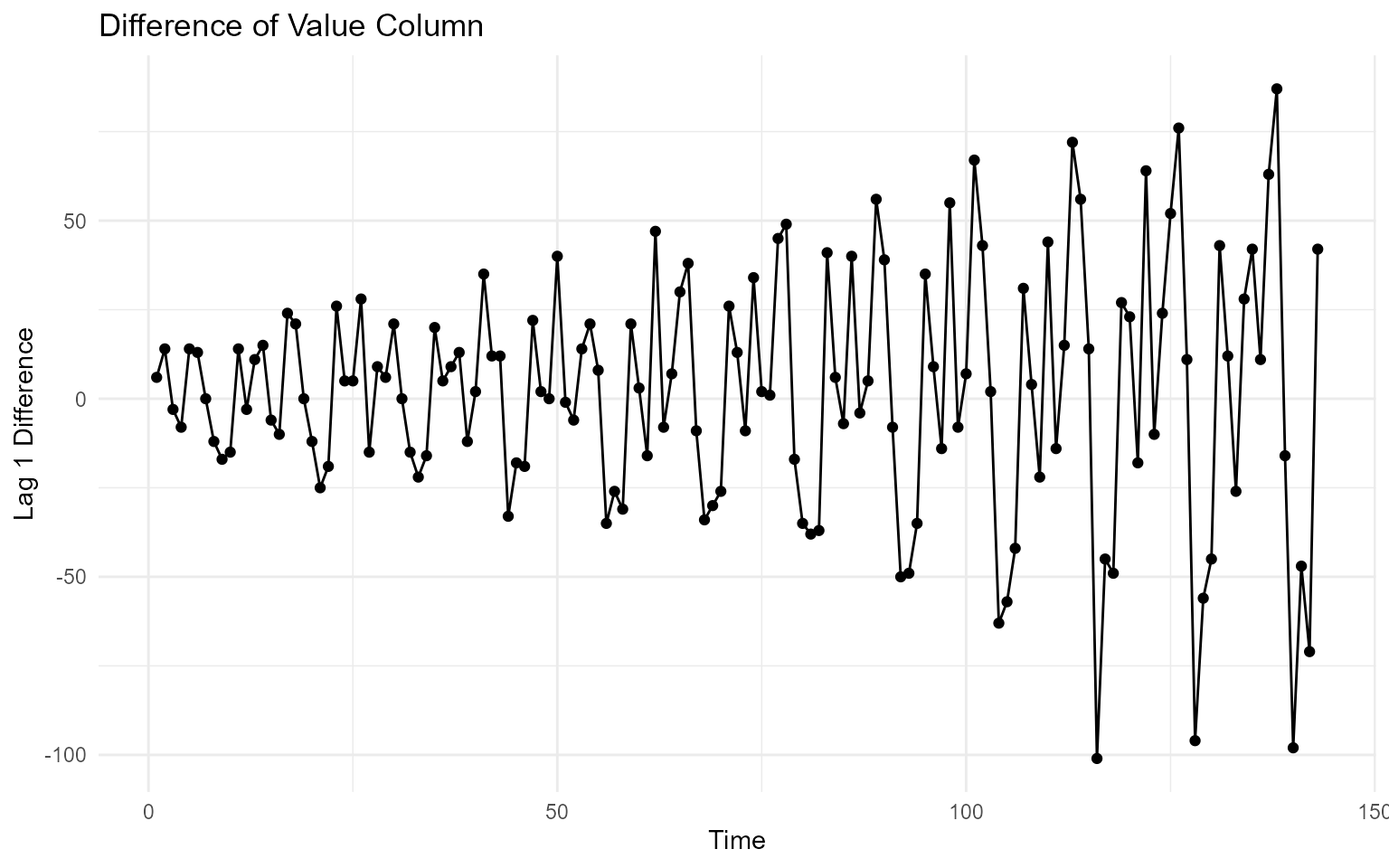
The differenced_value_tbl is a tibble that
has a lag 1 difference of the value column supplied.
dff_tbl
The dff_tbl is a tibble that is returned
that has the fft values, the complex, real and imaginary parts.
output$data$dff_tbl %>%
glimpse()
#> Rows: 144
#> Columns: 3
#> $ dff_trans <cpl> 40363.00000+0.00000i, 855.03235+8906.55958i, -48.11512+4098.…
#> $ real_part <dbl> 40363.00000, 855.03235, -48.11512, 517.59390, -137.07676, -2…
#> $ imag_part <dbl> 0.00000, 8906.55958, 4098.69669, 3225.75142, 2323.01117, 200…ts_obj
The last data piece of the data section is the ts_obj.
This is a ts version of the input_vector
output$data$ts_obj
#> Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
#> 1949 112 118 132 129 121 135 148 148 136 119 104 118
#> 1950 115 126 141 135 125 149 170 170 158 133 114 140
#> 1951 145 150 178 163 172 178 199 199 184 162 146 166
#> 1952 171 180 193 181 183 218 230 242 209 191 172 194
#> 1953 196 196 236 235 229 243 264 272 237 211 180 201
#> 1954 204 188 235 227 234 264 302 293 259 229 203 229
#> 1955 242 233 267 269 270 315 364 347 312 274 237 278
#> 1956 284 277 317 313 318 374 413 405 355 306 271 306
#> 1957 315 301 356 348 355 422 465 467 404 347 305 336
#> 1958 340 318 362 348 363 435 491 505 404 359 310 337
#> 1959 360 342 406 396 420 472 548 559 463 407 362 405
#> 1960 417 391 419 461 472 535 622 606 508 461 390 432Output Plots
There are a total of five plots that are returned in the list. Three
of them are ggplot plots and two of them are
plotly::ggplotly plots.
harmonic_plt
The harmonic_plot is a ggplot plot that
shows all of the harmonic waves on the same graph if you set
.harmonics greater than 1.
output$plots$harmonic_plot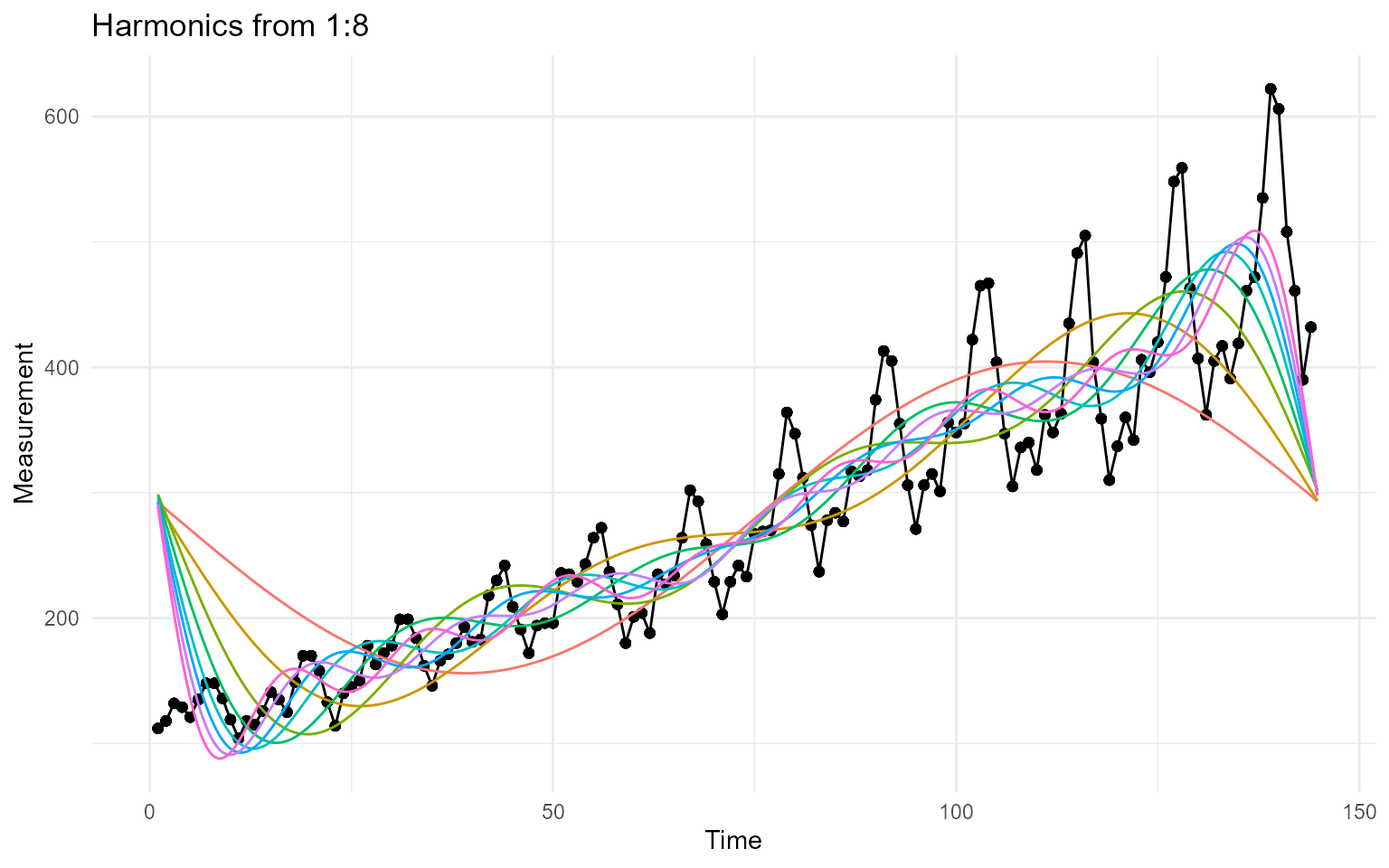
max_har_plot
The max_har_plot is a ggplot plot of the
maximum harmonic wave entered into .harmonics
output$plots$max_har_plot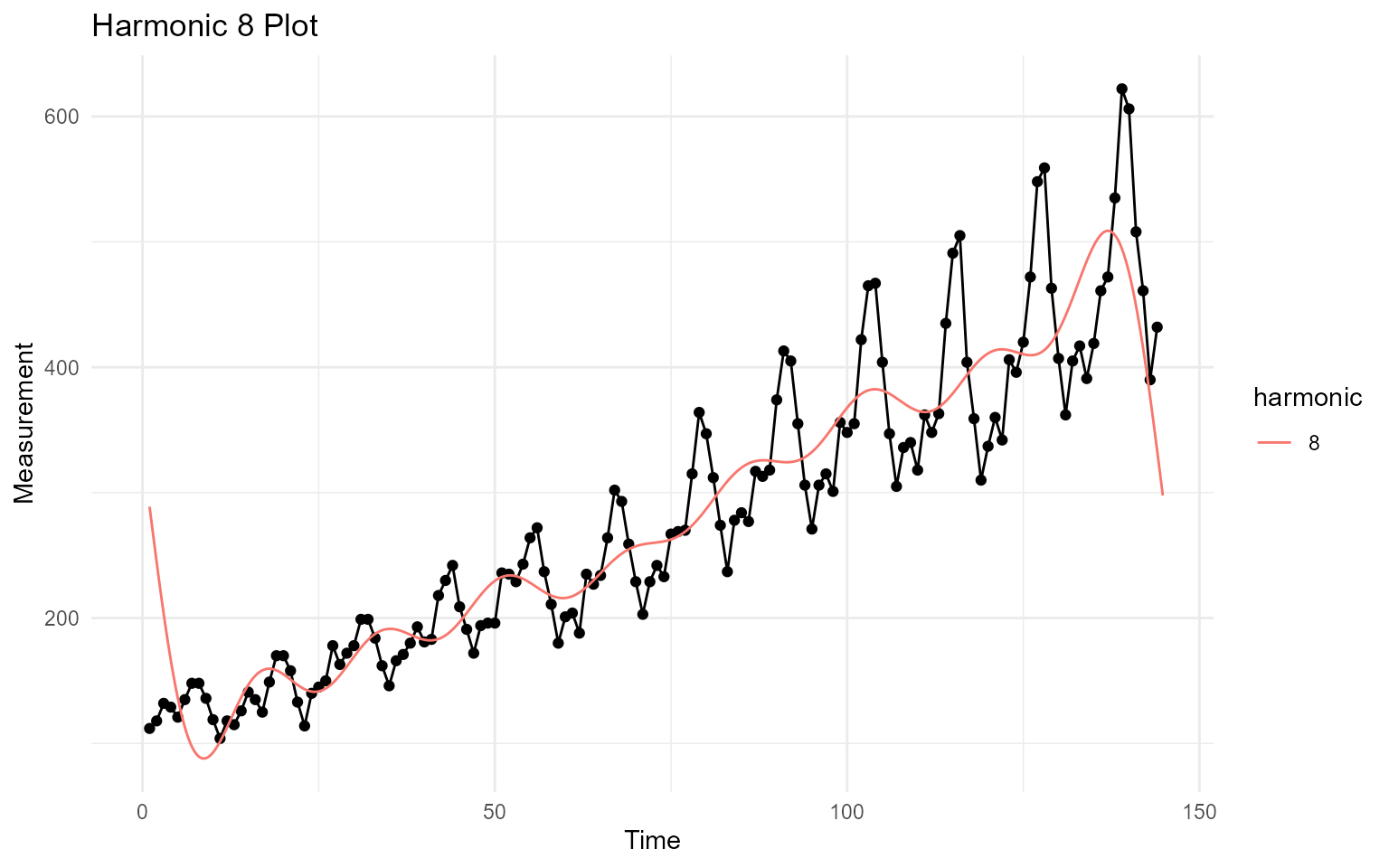
harmonic_plotly
The harmonic_plotly is a plotly::ggplotly
plot of the harmonic_plot
output$plots$harmonic_plotlymax_har_plotly
The max_har_plotly is a plotly::ggplotly
plot of the max_har_plot
output$plots$max_har_plotlyOutput Model
The model portion has four pieces to it which we will
look at below.
m
The parameter m is an internal parameter that is equal
to .harmonics / 2. This is fed into
TSA::harmonic along with the ts_obj
The parameter harmonic_obj is the object returned from
TSA::harmonic
The parameter harmonic_model is the harmonic model from
the TSA::harmonic
The parameter model_summary is a summary of the harmonic
model.
output$model$m
#> [1] 6
output$model$harmonic_obj %>% head()
#> cos(2*pi*t) cos(4*pi*t) cos(6*pi*t) cos(8*pi*t) cos(10*pi*t)
#> [1,] 1.000000e+00 1.0 1.000000e+00 1.0 1.000000e+00
#> [2,] 8.660254e-01 0.5 1.655735e-13 -0.5 -8.660254e-01
#> [3,] 5.000000e-01 -0.5 -1.000000e+00 -0.5 5.000000e-01
#> [4,] 1.157757e-12 -1.0 -5.292262e-12 1.0 3.969798e-12
#> [5,] -5.000000e-01 -0.5 1.000000e+00 -0.5 -5.000000e-01
#> [6,] -8.660254e-01 0.5 3.142992e-12 -0.5 8.660254e-01
#> cos(12*pi*t) sin(2*pi*t) sin(4*pi*t) sin(6*pi*t) sin(8*pi*t)
#> [1,] 1 -4.134027e-13 -8.268054e-13 2.397771e-12 -1.653611e-12
#> [2,] -1 5.000000e-01 8.660254e-01 1.000000e+00 8.660254e-01
#> [3,] 1 8.660254e-01 8.660254e-01 2.728918e-12 -8.660254e-01
#> [4,] -1 1.000000e+00 2.315515e-12 -1.000000e+00 -4.631030e-12
#> [5,] 1 8.660254e-01 -8.660254e-01 -5.796483e-13 8.660254e-01
#> [6,] -1 5.000000e-01 -8.660254e-01 1.000000e+00 -8.660254e-01
#> sin(10*pi*t)
#> [1,] -5.704992e-12
#> [2,] 5.000000e-01
#> [3,] -8.660254e-01
#> [4,] 1.000000e+00
#> [5,] -8.660254e-01
#> [6,] 5.000000e-01
output$model$harmonic_model
#>
#> Call:
#> stats::lm(formula = ts_obj ~ har_)
#>
#> Coefficients:
#> (Intercept) har_cos(2*pi*t) har_cos(4*pi*t) har_cos(6*pi*t)
#> 280.2986 -48.1494 16.7639 -6.3889
#> har_cos(8*pi*t) har_cos(10*pi*t) har_cos(12*pi*t) har_sin(2*pi*t)
#> 1.3889 -0.2534 -1.9097 -4.4632
#> har_sin(4*pi*t) har_sin(6*pi*t) har_sin(8*pi*t) har_sin(10*pi*t)
#> 11.6192 -11.1250 -7.9867 -6.4118
output$model$model_summary
#>
#> Call:
#> stats::lm(formula = ts_obj ~ har_)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -203.33 -93.48 -16.96 87.17 270.67
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 280.2986 9.8379 28.492 < 2e-16 ***
#> har_cos(2*pi*t) -48.1494 13.9128 -3.461 0.000726 ***
#> har_cos(4*pi*t) 16.7639 13.9128 1.205 0.230389
#> har_cos(6*pi*t) -6.3889 13.9128 -0.459 0.646840
#> har_cos(8*pi*t) 1.3889 13.9128 0.100 0.920632
#> har_cos(10*pi*t) -0.2534 13.9128 -0.018 0.985497
#> har_cos(12*pi*t) -1.9097 9.8379 -0.194 0.846381
#> har_sin(2*pi*t) -4.4632 13.9128 -0.321 0.748870
#> har_sin(4*pi*t) 11.6192 13.9128 0.835 0.405148
#> har_sin(6*pi*t) -11.1250 13.9128 -0.800 0.425367
#> har_sin(8*pi*t) -7.9867 13.9128 -0.574 0.566910
#> har_sin(10*pi*t) -6.4118 13.9128 -0.461 0.645662
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 118.1 on 132 degrees of freedom
#> Multiple R-squared: 0.1061, Adjusted R-squared: 0.03162
#> F-statistic: 1.424 on 11 and 132 DF, p-value: 0.169